1. Using itk-elastix: A first registration#
In this notebook we’ll learn the basics of how to register and transform images using itk-elastix.
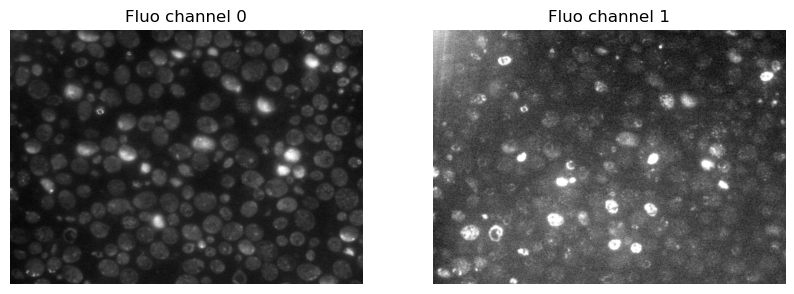
As example data, we’ll use two channels of a fluorescent image.
Also check the elastix manual.
import numpy as np
import pandas as pd
import matplotlib.pyplot as plt
Loading the data#
from skimage import io
im1 = io.imread("../example_data/fluo_ch0.tif")
im2 = io.imread("../example_data/fluo_ch1.tif")
# show the images next to each other
fig, ax = plt.subplots(1, 2, figsize=(10, 5))
ax[0].imshow(im1, cmap='gray')
ax[0].set_title('Fluo channel 0')
ax[0].axis('off')
ax[1].imshow(im2, cmap='gray')
ax[1].set_title('Fluo channel 1')
ax[1].axis('off')
plt.show()
Registration#
Importing itk-elastix#
Actually, the elastix functionality (relating to elastix and transformix) is contained with the Python package named itk, which was installed under the name itk-elastix.
import itk
Defining fixed and moving image#
Image registration typically works with a fixed and a moving image. The goal is to find the spatial transformation that makes the moving image align with the fixed image.
How to choose which image is fixed and which is moving? The fixed image is the one used as a reference, while the moving image is typically transformed to align with the fixed image.
fixed_image = im1
moving_image = im2
Defining image properties#
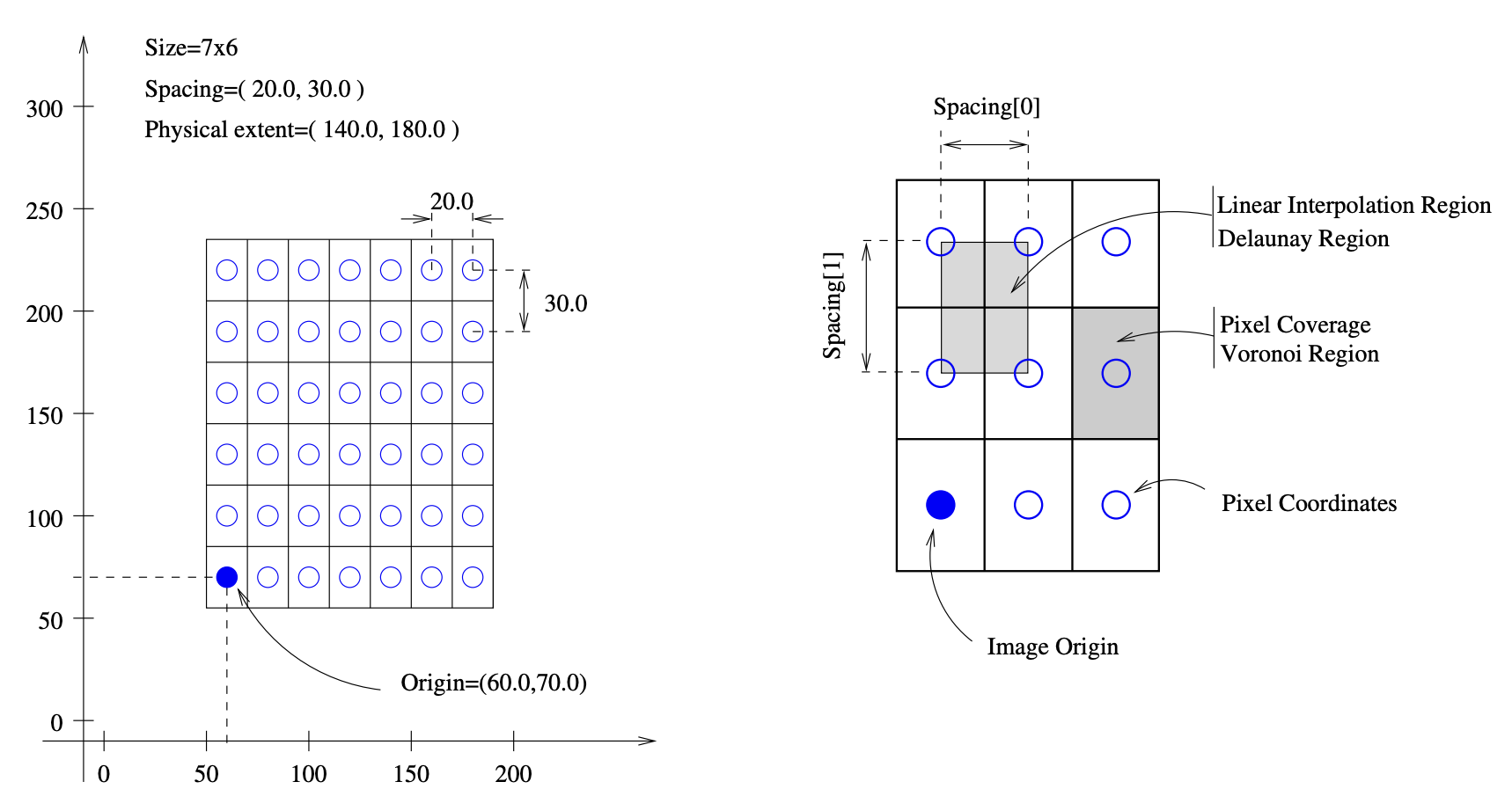
In the context of image registration it is important that image are placed into a physical coordinate system. For this purpose, we convert images originally loaded as numpy arrays into itk images. These have two important properties:
image spacing: the pixel spacing or physical distance between two images
image origin: a translational offset each images can have with respect to the origin of the coordinate system
If not set, default values of (1, 1) and (0, 0) are assumed.
pixel_spacing = [2, 2]
fixed_image_itk = itk.GetImageFromArray(im1)
moving_image_itk = itk.GetImageFromArray(im2)
# these lines are optional
fixed_image_itk.SetSpacing(tuple([float(v) for v in pixel_spacing])) # needs to be set as tuple of floats
fixed_image_itk.SetOrigin(tuple([float(v) for v in [0, 0]])) # needs to be set as tuple of floats
moving_image_itk.SetSpacing(tuple([float(v) for v in pixel_spacing])) # needs to be set as tuple of floats
moving_image_itk.SetOrigin(tuple([float(v) for v in [0, 0]])) # needs to be set as tuple of floats
Defining the registration parameters#
In command line elastix, the registration parameters are defined in a parameter file. In Python, we can define the parameters in a dictionary.
itk-elastix provides a function to create a default parameter map for a given registration task. We can then modify this parameter map to suit our needs.
# we can load the default parameter map for different types of transforms
# here we load the default parameter map for translation transforms
default_translation_parameter_map = itk.ParameterObject.New().GetDefaultParameterMap('translation')
# print the default parameter map
pd.DataFrame(
{'Parameters': default_translation_parameter_map.keys(),
'Values': default_translation_parameter_map.values()}
)
| Parameters | Values | |
|---|---|---|
| 0 | AutomaticParameterEstimation | (true,) |
| 1 | AutomaticTransformInitialization | (true,) |
| 2 | CheckNumberOfSamples | (true,) |
| 3 | DefaultPixelValue | (0,) |
| 4 | FinalBSplineInterpolationOrder | (3,) |
| 5 | FixedImagePyramid | (FixedSmoothingImagePyramid,) |
| 6 | ImageSampler | (RandomCoordinate,) |
| 7 | Interpolator | (LinearInterpolator,) |
| 8 | MaximumNumberOfIterations | (256,) |
| 9 | MaximumNumberOfSamplingAttempts | (8,) |
| 10 | Metric | (AdvancedMattesMutualInformation,) |
| 11 | MovingImagePyramid | (MovingSmoothingImagePyramid,) |
| 12 | NewSamplesEveryIteration | (true,) |
| 13 | NumberOfResolutions | (4,) |
| 14 | NumberOfSamplesForExactGradient | (4096,) |
| 15 | NumberOfSpatialSamples | (2048,) |
| 16 | Optimizer | (AdaptiveStochasticGradientDescent,) |
| 17 | Registration | (MultiResolutionRegistration,) |
| 18 | ResampleInterpolator | (FinalBSplineInterpolator,) |
| 19 | Resampler | (DefaultResampler,) |
| 20 | ResultImageFormat | (nii,) |
| 21 | Transform | (TranslationTransform,) |
| 22 | WriteIterationInfo | (false,) |
| 23 | WriteResultImage | (true,) |
# we could change a paremeter like in the following example
# notice that all parameter values are tuples of strings
default_translation_parameter_map['MaximumNumberOfIterations'] = ("300", )
# print the default parameter map
pd.DataFrame(
{'Parameters': default_translation_parameter_map.keys(),
'Values': default_translation_parameter_map.values()}
)
| Parameters | Values | |
|---|---|---|
| 0 | AutomaticParameterEstimation | (true,) |
| 1 | AutomaticTransformInitialization | (true,) |
| 2 | CheckNumberOfSamples | (true,) |
| 3 | DefaultPixelValue | (0,) |
| 4 | FinalBSplineInterpolationOrder | (3,) |
| 5 | FixedImagePyramid | (FixedSmoothingImagePyramid,) |
| 6 | ImageSampler | (RandomCoordinate,) |
| 7 | Interpolator | (LinearInterpolator,) |
| 8 | MaximumNumberOfIterations | (300,) |
| 9 | MaximumNumberOfSamplingAttempts | (8,) |
| 10 | Metric | (AdvancedMattesMutualInformation,) |
| 11 | MovingImagePyramid | (MovingSmoothingImagePyramid,) |
| 12 | NewSamplesEveryIteration | (true,) |
| 13 | NumberOfResolutions | (4,) |
| 14 | NumberOfSamplesForExactGradient | (4096,) |
| 15 | NumberOfSpatialSamples | (2048,) |
| 16 | Optimizer | (AdaptiveStochasticGradientDescent,) |
| 17 | Registration | (MultiResolutionRegistration,) |
| 18 | ResampleInterpolator | (FinalBSplineInterpolator,) |
| 19 | Resampler | (DefaultResampler,) |
| 20 | ResultImageFormat | (nii,) |
| 21 | Transform | (TranslationTransform,) |
| 22 | WriteIterationInfo | (false,) |
| 23 | WriteResultImage | (true,) |
# before running the registration, we need to create a
# registration parameter object to which we add the parameter map we just created
# (we could also add multiple parameter maps)
registration_parameter_object = itk.ParameterObject.New()
registration_parameter_object.AddParameterMap(default_translation_parameter_map)
Running the registration#
transformed_moving_image, result_transform_parameters = itk.elastix_registration_method(
fixed_image_itk,
moving_image_itk,
parameter_object=registration_parameter_object,
log_to_console=False
)
Visual inspection of the registration result#
transformed_moving_image is an itk image (which is an array + some metadata attached). However, it behaves almost like a numpy arrays.
For converting it into a numpy array, we can call itk.GetArrayFromImage(transformed_moving_image).
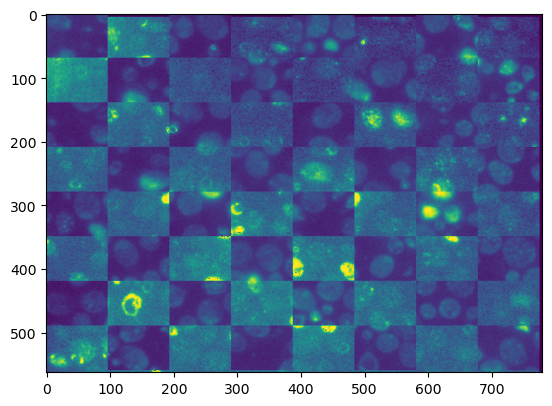
Option 1: Using a checkerboard pattern#
from skimage.util import compare_images
plt.imshow(compare_images(fixed_image_itk, transformed_moving_image, method='checkerboard'))
<matplotlib.image.AxesImage at 0x2824a44d0>
Option 2: Use a slider#
import jupyter_compare_view
Jupyter compare_view v0.2.4
%%compare
# overlay images in different colors
plt.figure()
plt.imshow(fixed_image, cmap='Blues')
plt.figure()
plt.imshow(transformed_moving_image, cmap='Reds', alpha=0.5)
Option 3: Use the napari viewer#
napari is very convenient for visualizing registration results because images are shown in world coordinates.
import napari
def add_itk_image_to_viewer(viewer, image_itk, layer_name, colormap):
viewer.add_image(
image_itk,
translate=fixed_image_itk.GetOrigin(),
scale=fixed_image_itk.GetSpacing(),
name=layer_name, colormap=colormap, blending='additive')
viewer = napari.Viewer()
add_itk_image_to_viewer(viewer, fixed_image_itk, 'fixed', 'Blue')
add_itk_image_to_viewer(viewer, moving_image_itk, 'moving', 'Red')
add_itk_image_to_viewer(viewer, transformed_moving_image, 'moving_transformed', 'Red')
Option 4: Load in Fiji#
save_dir = '../example_data/registration_output'
import os
if not os.path.exists(save_dir):
os.makedirs(save_dir)
io.imsave(os.path.join(save_dir, 'transformed_moving_image.tif'), transformed_moving_image)
Obtaining the resulting transformation#
Similarly to how we provided a registration_parameter_object, we obtain the resulting transformation in a transform_parameter_object.
We can extract the transformation parameters from this object.
transform_parameter_map = result_transform_parameters.GetParameterMap(0)
pd.DataFrame(
{'Parameter': transform_parameter_map.keys(),
'Value': transform_parameter_map.values()}
)
| Parameter | Value | |
|---|---|---|
| 0 | CompressResultImage | (false,) |
| 1 | DefaultPixelValue | (0,) |
| 2 | Direction | (1, 0, 0, 1) |
| 3 | FinalBSplineInterpolationOrder | (3,) |
| 4 | FixedImageDimension | (2,) |
| 5 | FixedInternalImagePixelType | (float,) |
| 6 | HowToCombineTransforms | (Compose,) |
| 7 | Index | (0, 0) |
| 8 | InitialTransformParameterFileName | (NoInitialTransform,) |
| 9 | MovingImageDimension | (2,) |
| 10 | MovingInternalImagePixelType | (float,) |
| 11 | NumberOfParameters | (2,) |
| 12 | Origin | (0, 0) |
| 13 | ResampleInterpolator | (FinalBSplineInterpolator,) |
| 14 | Resampler | (DefaultResampler,) |
| 15 | ResultImageFormat | (nii,) |
| 16 | ResultImagePixelType | (unsigned char,) |
| 17 | Size | (780, 563) |
| 18 | Spacing | (2, 2) |
| 19 | Transform | (TranslationTransform,) |
| 20 | TransformParameters | (8.15587732988515, -12.874474293169026) |
| 21 | UseDirectionCosines | (true,) |
Parameter interpretation#
Remember transform parameters are given in physical coordinates. They transform coordinates of the fixed to the moving image.


(illustration taken from https://www.orfeo-toolbox.org/SoftwareGuide/SoftwareGuidech9.html)
print('The obtained translational shift between the images (in physical units) is ',
[float(v) for v in transform_parameter_map['TransformParameters']])
The obtained translational shift between the images (in physical units) is [8.15587732988515, -12.874474293169026]
Transform an image#
Sometimes it’s useful to apply the resulting transform parameters to an image independently of a registration run. Or to a different image.
In command line elastix, we’d use the transformix tool for this.
# apply the obtained transform parameters without previously performing registration again
result_image_transformix = itk.transformix_filter(
moving_image,
result_transform_parameters)
%%compare
plt.figure()
plt.imshow(transformed_moving_image, cmap='Blues')
plt.figure()
plt.imshow(result_image_transformix, cmap='Reds', alpha=0.5)
What went wrong? Shouldn’t the two images above be the same?#
The registration parameters are defined in physical space, so we need to use the itk image with the correct spacing and origin for performing the registration.
result_image_transformix = itk.transformix_filter(
moving_image_itk, # notice the itk here
result_transform_parameters)
%%compare
plt.figure()
plt.imshow(transformed_moving_image, cmap='Blues')
plt.figure()
plt.imshow(result_image_transformix, cmap='Reds', alpha=0.5)